Genomics: Insight

Can Genetically Engineered Bacteria Solve The Plastic Pollution Crisis?
Tackling Plastic Waste with Plastic-Eating Bacterial Enzymes
Plastic is an innovation which has pervaded every aspect of modern life, but human dependence on plastic comes at a heavy price. Plastic remains in the environment for millenia, breaks down into microplastics which find their way into our bodies, and gravely threaten the health of humanity and our environment. However, genomics and biotechnology hold the key to solving this pressing problem. As the name suggests, plastic-eating bacteria are recently discovered organisms which are capable of digesting man-made plastics. Coupled with genomic analysis and genetic engineering, these microbes have the potential of producing a revolutionary solution to the plastic pollution crisis.
What are plastic eating bacteria?
Plastic-eating bacteria are a classification of microorganisms which can digest synthetic polymers. Plastic cannot be efficiently broken down and remains in the environment for millennia because plastic has only been mass produced in the past century and decomposers have not adapted quickly enough to digest its complex molecular structure. However, the discovery of plastic-eating bacteria negates the previous belief that plastic could not be naturally digested by decomposers. The abilities of plastic-eating bacteria and other organisms are very unique from a genomics and evolutionary context as they demonstrate the emergence of a new behavior which did not exist before. However, their ability to consume artificial polymers are due to the various enzymes produced in their digestive systems.
“Plastic-eating bacteria are a classification of microorganisms which can digest synthetic polymers.”
What types of plastic-eating bacteria and plastic-eating enzymes are there?
Plastic-eating bacteria can be characterized into two groups - nylon and PET (two different types of plastic) eating bacteria. The first type was discovered in 1975 from the waste water drainage ponds of a Japanese nylon plant. Then named Flavobacterium sp. K172 did not exist before the mass production of nylon and was found to survive solely on cyclic dimers like 6-aminohexanoic acid, a byproduct of Nylon-6 production. They also discovered that 6-aminohexanoic acid cyclic dimer hydrolase and 6-aminohexanoic acid linear oligomer hydrolase, the enzymes responsible for nylon digestion could not digest any other compound except artificial nylon byproducts.3 Collectively, these bacterial digestive enzymes became known as nylonases. Later studies suggest that the gene nylonases like 6-aminohexanoic acid hydrolase arose from the combination of a gene duplication and a random frameshift mutation and that these changes in multiple genes contributed to the evolution of the strain. 4
The other form of plastic-eating bacteria are PET-eating bacteria. The first strain of PET-eating bacteria was isolated in 2016 from a plant in Sakai City, Japan. Named Ideonella sakaiensis, the gram-negative, rod-shaped bacteria, was found to be closely related to the non-plastic eating species of Ideonella dechloratans and Ideonella azotifigens through RNA gene sequencing and phylogenetic analysis.5 PET-eating bacteria use two enzymes to digest PET - PETase and MHETase. The bacteria secretes both enzymes simultaneously which break down PET into environmentally benign substances. A colony of I. sakaiensis can completely degrade a low-grade plastic water bottle in six weeks. Higher-grade PET products would require heating and cooling to weaken it before bacteria could start eating.7
Independent enzymes capable of digesting PET have also been discovered. In 2012, researchers found leaf-branch compost cutinase (LLC). This enzyme evolved to break down the waxy protective coating on many plants’ leaves but adapted to PET’s chemical bonds. However, the enzyme can only function after days at a specific temperature, where it slowly begins breaking down the polymer bonds.10
How do nylonases and PETases work?
The mechanism behind all plastic-digesting bacteria and enzymes is the severing of the chemical bonds, or esters, in a synthetic polymer like nylon or PET. This breaks down the complex molecule into simpler monomers which this organism can then digest. The nylonase NylB, encoded by the nylB gene, can only degrade nylon dimers and short oligomers. This means that nylonase can only digest nylon waste byproducts.
PET has a crystalline structure, so it is recalcitrant to catalytic depolymerization as the ester linkages are too strong. PETase works by converting PET to mono(2-hydroxyethyl) terephthalic acid (MHET), with trace amounts of terephthalic acid (TPA) and bis(2-hydroxyethyl)-TPA as secondary products. MHETase works hand in hand with PETase, further converting the MHET into its two monomers, TPA and ethylene glycol (EG) which are then digestible by I. sakaiensis and serve as an energy source.
Like PETase in bacteria, LLC works by snipping the bonds between PET’s two building blocks: terephthalate and ethylene glycol. But LLC can only wiggle into the polymer linkages and breaks them down after a few days of working at 65°C, the temperature at which PET softens, after which it naturally denatures. (Fig. 1)
How can we use plastic eating enzymes and gene editing to combat plastic pollution?
Scientists were able to induce other species of bacteria like Pseudomonas aeruginosa and E. coli, to evolve the capability to break down the same nylon byproducts of nylonase in a laboratory by forcing them to live in an environment with no other source of nutrients and plasmid transfer.4 These examples show that nylonase can be harnessed to be used in a variety of inexpensive and easily accessible species of bacteria which can then be used en mass to tackle plastic pollution.
P. aeruginosa can also digest PET and other plastics like polyethylene, polypropylene, and many more with varying degrees of efficiency. PETase also has attributes which make it a plausible method of combating plastic - it is physically tailored to bind to PET surfaces and works at 30°C, making it suitable for recycling in bioreactors.
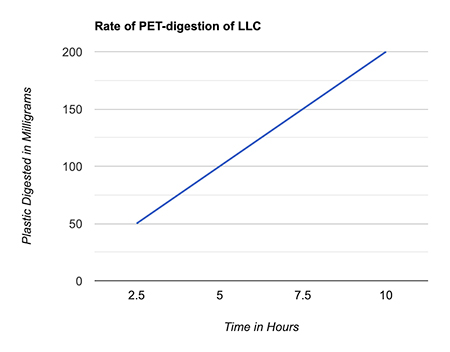 Figure 1. The rate at which the enzyme LLC consumes PET in magnitude of milligrams per hour (created by author JiaJia Fu using Google Sheets).
Figure 1. The rate at which the enzyme LLC consumes PET in magnitude of milligrams per hour (created by author JiaJia Fu using Google Sheets).Why are plastic-eating enzymes better than existing solutions for managing plastic waste?
Only 9% of all plastics produced are recycled due to the high cost of chemically breaking down plastic relative to its cheap and efficient production costs.1 As recycling is expensive and resource consuming, the majority of plastic goes to landfills or the environment. Most proposed solutions for plastic waste only target the aftermath - when plastic enters the environment and becomes pollution. Other than attempting to reduce plastic production and its entering the environment (physical measures like nets to collect plastic in the ocean), humans have not found a way of actually eliminating the waste itself until now. Gene-editing using CRISPR and other genomics techniques also hold the key to producing a viable and affordable solution by utilizing natural mechanisms. Even if a specific bacterial strain or enzyme can only digest one type of plastic or breaks down its respective constituent too slowly, situations which are both inapplicable in a commercial context, researchers can alter them to make the process faster, more efficient, and effective. Through analyzing the crystal structure of the enzyme and identifying key amino acids, researchers isolated a mutant enzyme 10,000 times more efficient at PET breaking than natural LLC. The team found that it could break down 90% of 200 grams of PET in 10 hours. They converted 90% of that same plastic back to its starting materials, and resynthesized PET out of the terephthalate and ethylene glycol composites - the recycled PET was just as strong as conventional plastic. The success of their trials has led to scaling up the technology and plans to open a demonstration plant next year.
“Most proposed solutions for plastic waste only target the aftermath - when plastic enters the environment and becomes pollution.”
What are potential impacts and consequences of plastic-eating enzymes?
Thanks to genomics and biotechnology, the potential of a unique biological mechanism can be fully realized and even begin to be applied on a massive scale. The impacts and application of a technology or process developed from plastic-eating bacteria and enzymes is immeasurable. Plastic-eating enzymes could bring about a reliable industrial plastic waste management process, truly effective recycling, ocean plastic and microplastic eliminating serums, or even at home, plastic waste consuming compost bins.
The potential impacts of utilizing plastic-eating bacteria are undeniable, but so are the possible setbacks and consequences of deploying this technology.
Although genetic manipulation can unlock the potential of nylonase, at present it cannot be utilized on a mass scale as they cannot digest commercial nylon, but waste product nylons which have already been degraded into short oligomers. The economic viability of LLC also remains unclear. It can only break down pure PET and not dyes and other plastics mixed in plastic products. Introducing gene edited life into the general environment could have unforeseen consequences. There’s no way to predict how a plastic-eating bacteria by itself, not even considering genetic manipulation, would react with a new ecosystem like the ocean, or a landfill. The ways bacteria could mutate and interact with its environment are too unpredictable to release without proper research and considerations.
Although there are concerns and risks involved with developing such a technology, unquestionably plastic-eating bacterial enzymes may lead the efforts to quell the environmental disaster the world is facing. Through utilizing genomics, cutting edge biology, and nature, we are one step closer to solving the plastic pollution crisis and forging a healthier society, planet, and future.
“Through utilizing genomics, cutting edge biology, and nature, we are one step closer to solving the plastic pollution crisis and forging a healthier society, planet, and future.”
About the Author
JiaJia Fu is a freshman student at Whittle School and Studios. Born and raised in New Jersey, she is passionate about science and nature and aspires to create breakthrough solutions for ecological problems like plastic pollution, intensive agriculture, and climate change through biotechnology.