Genomics: Insight
Genomics and the Fight Against Artemisinin Resistance
Using Genomic Epidemiology to Combat the Spread of Drug Resistant Malaria Parasites

Malaria is one of the most significant public health problems worldwide. An estimated 228 million cases occurred in 2018, causing approximately 405,000 deaths1. Malaria is a mosquito-borne infectious disease that causes fever, tiredness, anemia, vomiting, headaches, and other symptoms. In severe cases, coma, organ failure, or death may occur. Malaria is caused by parasites of the species Plasmodium. The majority of clinically severe cases are caused by the species Plasmodium falciparum. The global incidence of malaria declined from 2010 to 2018, but in recent years, the rate of that decline has slowed substantially, indicating that progress in eliminating the disease is slowing1.
Artemisinin-based combination therapies (ACTs) have contributed to recent reductions in morbidity and mortality. Artemisinin derivatives were discovered in the 1970s and are potent antimalarials. Because they possess short half-lives, they are used in tandem with longer-acting drugs, including amodiaquine, piperaquine, mefloquine, sulfadoxine/pyrimethamine, or lumefantrine. These efficacious combination therapies have become a cornerstone of malaria treatment in endemic areas. The use of ACTs has been responsible for material reductions in parasite burdens and deaths in Africa2.
Different anti-malarial drugs work through a variety of mechanisms, including blocking parasite detoxification pathways, interfering with folate metabolism, and inhibiting mitochondrial function. Artemisinin derivatives have a complex mechanism of action that can kill the parasite at a variety of life cycle stages by causing widespread damage to parasite proteins. Artemisinin and its derivatives are activated via exposure to free heme, an iron containing compound that composes the non-protein portion of hemoglobin. Exposure to heme results in cleavage into artemisinin’s active form, dihydroartemisinin (DHA). DHA then causes irreversible damage to parasite proteins and the accumulation of these unfolded and damaged proteins. Build-up of damaged proteins activates the endoplasmic reticulum’s stress response and leads to parasite death3.
Increasing prevalence of artemisinin resistant P.
falciparum across large parts of Southeast Asia threatens to destabilize malaria control worldwide
Plasmodium drug resistance, the ability of parasites to survive and reproduce despite the administration of antimalarials, is a recurring problem in control and elimination efforts. Resistance has emerged to all known antimalarials, and emerging resistant parasites are a significant threat to progress against malaria. The recent emergence of resistance to ACTs in Southeast Asia is of particular concern. Some parasites are now able to tolerate artemisinin in their young life stages by entering a period of dormancy and then resuming growth after drug treatment has subsided, resulting in lower rates of parasite clearance4. Evidence of artemisinin resistance was first noted in 2008 in Cambodia, and resistant parasites are now spreading throughout the Greater Mekong Region5. For example, dihydroartemisinin-piperaquine treatment had failure rates between 14.3% and 46.3% in studies conducted from 2015-2017 in Vietnam and artesunate-mefloquine replaced dihydroartemisinin- piperaquine as the recommended first line treatment in Cambodia after high failure rates were observed6. The spread of resistance to Africa from Southeast Asia via Bangladesh and India is of concern because this path was the source of the now-pervasive resistance to another antimalarial, chloroquine, in Africa7. Resistance to ACTs in Africa could result in substantial epidemics and jeopardize recent progress8. Preventing the establishment of ACTs resistance in Africa represents a challenge to current malaria control efforts.
Decreased artemisinin efficacy in young parasites is associated with mutations in Kelch13, a protein located on chromosome 13. The precise mechanism of this resistance is not known, but mutations in Kelch13 could help parasites resist injury from artemisinin by protecting proteins from damage, preventing the accumulation of damaged proteins, or decreasing the amount of free heme and
reducing the activation of artemisinin itself. Additionally, Kelch13 mutations could delay parasite growth, allowing the parasite to mimic a dormant phase and resist damage4. Kelch13 was identified as a molecular marker for artemisinin resistance in 20149. Initial identification of molecular resistance markers, like mutations in Kelch13, involves the use of genome-wide association studies (GWAS) and other genetic association studies to (1) find positions on the chromosome that are under strong selective pressure; and (2) identify specific mutations associated with resistance2,10. Once a marker is identified, it must be validated via a variety of laboratory studies. More than 20 different single nucleotide polymorphisms (SNPs) in Kelch13 that confer artemisinin resistance have now been identified, and these polymorphisms have resulted from independent mutational events10,11.
Genomic data such as whole genome sequencing (WGS) data can be used to track the prevalence and spread of these resistant strains by providing detailed information on parasite population structure. A study from the MalariaGEN (Malaria Genomic Epidemiology Network) P. falciparum Community Project analyzed sequencing data from more 3,000 clinical samples from 23 countries in Africa and Southeast Asia. The study determined that kelch13 mutations appear infrequently in Africa and have occurred independently from Southeast Asian parasites. These mutations are maintained at low levels in the population and have not undergone the same degree of selective pressure as those in Southeast Asia, demonstrating that factors beyond mutation alone affect the spread of resistance. Patterns of drug treatments and other environmental factors also determine how mutations spread, and genomic data is an essential tool in trying to disentangle these complex systems. These findings demonstrate the importance of applying genomic data to understanding why resistance spreads and ensuring that ACT-resistant malaria does not become a significant issue in Africa12.
Another recent study analyzed more than 2,000 genomes from the MalariaGEN P. falciparum Community Project collected between 2007 and 2018 to gather fine-scale epidemiological data on the KEL1/PLA1 strain of parasite that is resistant to dihydroartemisinin-piperaquine treatment. KEL1/PLA1 parasites have spread across the Eastern Southeast Asia since 2015, replacing many of the previous local parasite populations. Multiple groups of these parasites have spread in independent waves. It is likely that exposure to dihydroartemisinin-piperaquine after resistance emerged created increased drug pressure and resulted in parasite populations with further mutations that are more extensively resistant to treatment13. This study highlights the importance of genetic data in informing surveillance efforts and preventing the emergence of multidrug-resistant species.
To date, most genomic and molecular surveillance techniques have been employed primarily in research contexts and have not been fully integrated into routine surveillance practices14. Recent advances in sequencing technologies and reductions in associated costs are making genomic techniques more accessible. The cost of DNA sequencing per raw megabase, 1 million nucleotides, has declined from $10,000,000 in 2001 to $0.01 in 2019 with the steepest declines in cost occurring after 2008 when many centers transitioned to next-generation sequencing from traditional Sanger sequencing15. The ability to obtain WGS from dried blood spot (DBS) where blood samples are blotted and dried on filter paper was a significant breakthrough in translating genomic technology to real world malaria surveillance efforts. DBSs can be acquired with minimal resources and personnel and can be stored without refrigeration, making them an ideal form of sample collection in resource-limited settings16. While sequencing techniques still require investment in infrastructure, computing power, and trained personnel, the development of centralized national and sub-regional reference laboratories could help
The integration of epidemiological, molecular, and genomic data could substantially improve malaria surveillance in elimination settings
reduce these costs. Additionally, WGS is not needed in all contexts. Once new genetic variants have been identified and molecular markers are validated, lower-cost methods that target specific genome regions can be used. Data and sample sharing is also essential. Initiatives like the MalariaGEN have provided examples of successful collaborative efforts14. There is still time to prevent the spread of ACT resistance on a global scale, but these efforts will require the efficient integration of epidemiological, molecular, and genomic data into routine surveillance.
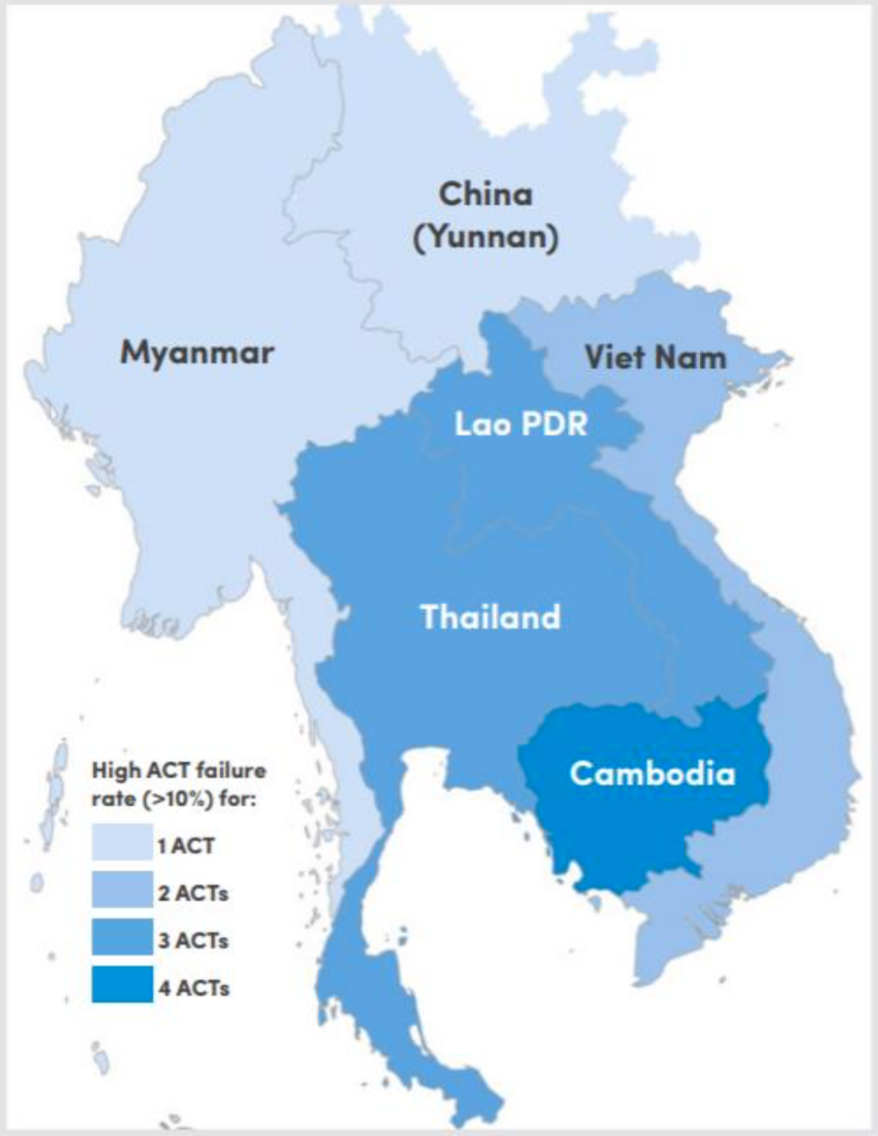
Figure 1. Classification of Southeast Asian countries by number of ACTs failing (>10% treatment failure) after 2010. (Source: WHO Status Report on Artemisinin Resistance and ACT Efficacy December 2018
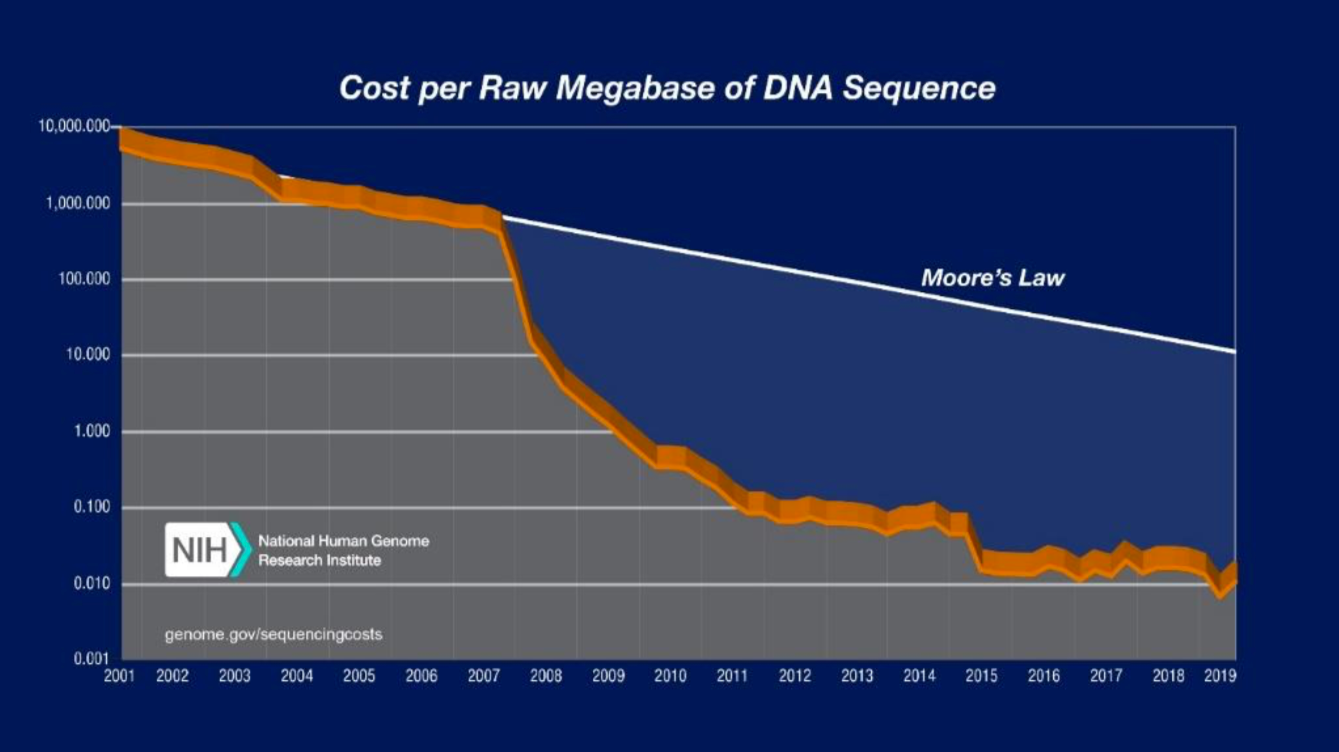
Figure 2. Decreasing Sequencing Costs from 2001 to 2019. The graph shows the costs associated with DNA sequencing from 2001 to 2019. Moore’s law describes a trend in the computer hardware industry where computing power doubles every two years. Technology that progresses on pace with Moore’s law is generally regarded as progressing on pace. Sequencing costs have declined rapidly, significantly outpacing Moore’s law. (Source: DNA Sequencing Costs: Data)
About the Author

Emma Rowley is a postbaccalaureate fellow in the National Institute of Allergy and Infectious Disease’s Laboratory of Malaria and Vector Research where she is researching Plasmodium vivax, a parasite that causes malaria. She graduated from Wake Forest University in 2019 with a Bachelor of Science in Biology.