Genomics: Insight

The Role of Non-Coding RNA in Potential Cancer Therapeutics
Introduction
The major underlying cause of oncogenesis, the formation of tumors, is dysregulated expression of certain genes in crucial growth regulatory pathways. Recently, research has shown that abnormal gene expression not only resulted from protein-coding genes, but also non-coding genomic elements [4]. In 2012, the Encyclopedia of DNA Elements transcriptome project (ENCODE) concluded that only 1.2% of the human genome codes for proteins, while about 80% are actively transcribed into different variations of non-coding RNAs (ncRNAs) [4]. NcRNAs are involved in gene regulation at different levels, from epigenetic gene silencing to post-transcriptional regulation of mRNA stability [3]. NcRNA can cause changes in cellular processes leading to increased cell proliferation by interacting with proteins, DNA, and RNA, which can indirectly contribute to histone modification, DNA methylation, and heterochromatin development (condensed chromosome formed during cell division) [3]. Additionally, ncRNA-based cancer drugs provide a better alternative than current therapeutics. The current therapeutic agents only target a single gene or pathway, and they are consequently only effective for a limited period of time until the cancer cells develop resistance. However, a single ncRNA can control the expression of many downstream mRNA targets and affect multiple interconnected growth regulatory pathways, thus preventing drug resistance by disabling the pathways cancer can utilize to reappear [6].
Abnormal gene expression … is also largely due to the regulation of non-coding genomic elements.
MiRNA-based cancer drugs in suppressing oncogenes
NcRNA can be roughly categorized into two groups based on size, small ncRNAs (sncRNAs), shorter than 200 nucleotides, and long ncRNAs (lncRNA), longer than 200 nucleotides [3]. The category of sncRNA can then be further broken down into subcategories, the most studied of which is microRNAs (miRNAs). The structure of miRNA allows it to target hundreds of transcripts, making them powerful regulators whose abnormal expression can have a significant impact on cancer development. Currently, the two most promising cancer therapies are miRNA mimics and miRNA inhibitors [6].
MiRNA mimics are double-stranded synthetic oligonucleotides that are processed to form active, single-stranded molecules upon entering cancer cells. Those fragments mimic the native miRNAs and bind to RNA-induced silencing complexes (RICS), which then bind to target mRNAs, resulting in post-transcriptional repression and preventing oncogenes from being translated [6]. However, the disadvantage of miRNA mimics is their instability. MiRNA mimics degrade quickly in biological fluids, thus requiring a unique protective delivery system. Nevertheless, several miRNA therapies have passed the initial litmus test and have proceeded to early-phase clinical testing, including MRX34, a miRNA mimics of tumor suppressor miR-34a which halts metastasis (secondary cancer) as well as stemness (cell self-renewal) [6].
Compared to miRNA mimics, miRNA inhibitors are easier to design due to their structures and pharmacokinetic properties (how drugs travel in the body). There are multiple methods to produce miRNA inhibitors [4]. The first method is the oligonucleotide-based approach. Antisense oligonucleotides (ASOs) are single-stranded RNAs complementary to the target mRNA sequences and act as competitive inhibitors to suppress target miRNAs. Similar to ASO, miRNA sponges contain complementary sequences to their target miRNAs, and each miRNA sponge can block multiple miRNAs with the same target binding sequence. However, miRNA sponges require a greater concentration to achieve the same effect as ASO, thus increasing the severity of side effects [6]. The third method to generate miRNA inhibitors is through CRISPR/Cas 9, which can cut genetic material with precision using single guide RNAs (sgRNAs). CRISPR can cut miRNA-producing regions in the DNA, leading to downregulation of mature miRNAs while minimizing the off-target effects. Reduced amounts of miRNAs can result in reduced cell proliferation and metastasis [7]. However, this method is not widely utilized in therapeutic settings because the current delivery system is not efficient at delivering CRISPR/Cas 9 machinery to living cells [6]. The figure below shows both miRNA mimics and miRNA inhibitors, as well as their individual pathways to degrading mRNAs, inactivating miRNAs, and directly changing DNA sequences.
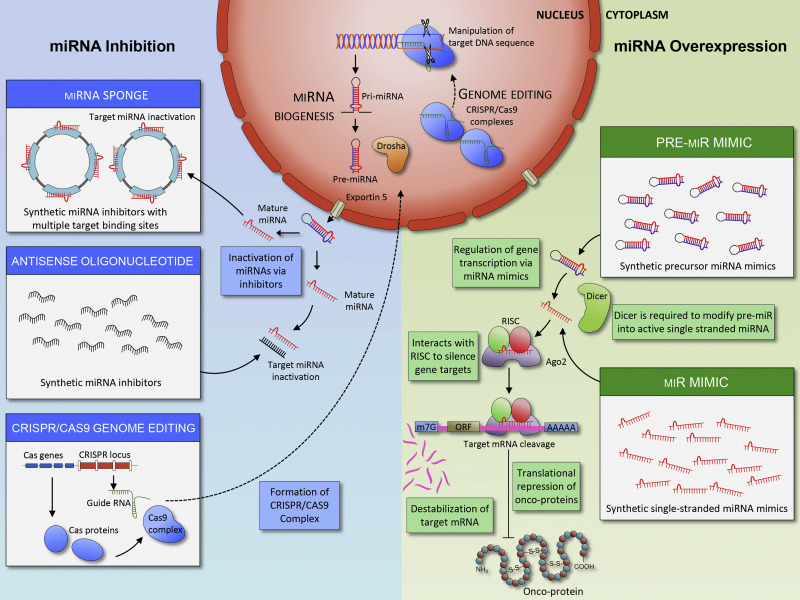 Figure 1: Mi-RNA based cancer therapeutics (Toden, Zumwalt, & Goel, 2021) On the right, the figure shows miRNA mimics, which prevent translation of oncoproteins by forming RISCs. On the left, the figure shows three ways miRNA inhibitors can be made and how they interact with miRNA and DNA.
Figure 1: Mi-RNA based cancer therapeutics (Toden, Zumwalt, & Goel, 2021) On the right, the figure shows miRNA mimics, which prevent translation of oncoproteins by forming RISCs. On the left, the figure shows three ways miRNA inhibitors can be made and how they interact with miRNA and DNA.The structure of miRNA allows it to target hundreds of transcripts, making them powerful regulators…
LncRNAs and their functions in the p53 pathway
The other group of ncRNAs, long ncRNA (lncRNAs) can fold into more complex three-dimensional shapes because of their longer structure. Unlike sncRNAs, lncRNAs can be tissue-specific, cell-specific, and even compartment-specific, making them the “fine tuners” of gene regulation [2]. Abnormal expression of lncRNA can result in cell proliferation, tumor progression, and metastasis. The bulk of the current research on lncRNAs is focused on the tumor suppressor p53, which triggers cell cycle arrest and apoptosis. Several lncRNAs are involved in the p53 transduction pathways, including the lncRNA activator of enhancer domains (LED) [5]. While p53 activates the transcription of LED, LED also contributes to the regulation of p53. Various cancers, including breast cancer and colorectal cancer, show downregulation of LED expression [5]. LED is essential to the p53 pathway, and artificially overexpressing LED in cancer cells could potentially lead to the activation of p53, making it a potential target in cancer therapy [1].
Another divergent transcription of p53 is the recently discovered lncRNA GUARDIN, which promotes cell survival and genome stability. The absence of GUARDIN results in telomere fusion, which activates apoptosis. For instance, in p53-mutant colorectal cancer, GUARDIN expression is significantly reduced. Overall, GUARDIN may be a potential novel target in p53-mutant tumors, yet further research on its possible ambiguous phenotypes in certain types of tumors is needed [5].
A unique class of ncRNAs: CircRNAs
Circular RNAs (circRNAs) are a unique class of ncRNAs. Unlike sncRNA and lncRNA whose structures are linear, circRNA contains covalently closed loops with neither 5’ and 3’ polarities nor polyadenylated tails. This particularly stable structure shows great potential as an efficient deliverable drug [6]. CircRNAs have specific binding sites, regulate mRNA splicing during post-transcription, and act as miRNA sponges or miRNA inhibitors. For example, ciRS-7 is a newly identified circRNA that functions as a miR-7 sponge. Research shows that the expression of ciRS-7 is completely absent in cancer cells [8]. However, in the tumor microenvironment, the system that surrounds the tumor, ciRS-7 is highly expressed [8]. This contrast demonstrates the complex role of circRNA as well as the need for further research.
Conclusion
Over the past two decades, a new forefront of cancer therapy using ncRNAs arose. Studies have extensively explored the role of ncRNAs in cancer progression and their participation in drug resistance. Nonetheless, numerous obstacles need to be conquered before human clinical trials can begin. NcRNA is still a relatively new subject, and there is much to learn about the many aspects of their biogenesis and function, as well as the appropriate methodology for the therapeutic delivery of these ncRNAs.
…there is much to learn about the many aspects of ncRNA’s biogenesis and function…
Related Links
- Lee, J. H., Xiong, F., & Li, W. (2020). Enhancer RNAs in cancer: regulation, mechanisms and therapeutic potential. RNA biology, 17(11), 1550–1559. https://doi.org/10.1080/15476286.2020.1712895
- Ling, H., Vincent, K., Pichler, M. et al. (2015). Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 34, 5003–5011. https://doi.org/10.1038/onc.2014.456
- Panni, S., Lovering. R., Porras, P., Orchard, S. (2020). Non-coding RNA regulatory networks. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, 1863(6), 194417. https://doi.org/10.1016/j.bbagrm.2019.194417
- Romano, G., Veneziano, D., Acunzo. M., Croce, C. (2017). Small non-coding RNA and cancer. Carcinogenesis, 38(5), 485–491. https://doi.org/10.1093/carcin/bgx026
- Sanchez Calle, A., Kawamura, Y., Yamamoto, Y., Takeshita, F., & Ochiya, T. (2018). Emerging roles of long non-coding RNA in cancer. Cancer science, 109(7), 2093–2100. https://doi.org/10.1111/cas.13642
- Toden, S., Zumwalt, T. J., & Goel, A. (2021). Non-coding RNAs and potential therapeutic targeting in cancer. Biochimica et biophysica acta. Reviews on cancer, 1875(1), 188491. https://doi.org/10.1016/j.bbcan.2020.188491
- Yang, J., Meng, X., Pan, J., Jiang, N., Zhou, C., Wu, Z., & Gong, Z. (2018). CRISPR/Cas9-mediated noncoding RNA editing in human cancers. RNA biology, 15(1), 35–43. https://doi.org/10.1080/15476286.2017.1391443
- Zhang, Z., Xie, Q., He, D., Ling, Y., Li, Y., Li, J., & Zhang, H. (2018). Circular RNA: new star, new hope in cancer. BMC cancer, 18(1), 834. https://doi.org/10.1186/s12885-018-4689-7
About the Author
Gloria Bao is a sophomore at Thomas Jefferson High School for Science and Technology. She is passionate about genetic engineering and computational biology as well as their applications in cancer therapeutics.