Genomics: Insight
Socioeconomic Status, Epigenetics, and Mental Health
Introduction
Epigenetics is the study of changes in gene expression without altering the DNA sequence itself. Changes in gene expression can result from DNA methylation, which is when an extra methyl group (a carbon and three hydrogen atoms) is added to a cytosine nucleotide (Wang & Chang, 2018). DNA methylation occurs at CpG sites which are long stretches of cytosine-guanine nucleotides that are typically involved in the regulation of a certain gene. Observing changes in methylation of these sites is useful for making associations between particular genes and environmental factors (Long et al., 2016). Environmental factors, such as stress, can influence epigenetic changes which in turn can alter the expression of genes. This can impact genes that affect mental health and cause adverse mental health outcomes. Socioeconomic status (SES) is used to define a person’s class or standing within society. Typically, occupation, education, and income are factors used to determine how high or low a person’s SES is (American Public Health Association, 2015). SES can be measured at the level of the individual or with the context of their community. Health is commonly affected by what an individual’s SES is, and having a low SES has been shown to put a person more at risk of experiencing adverse health outcomes (Schultz et al., 2018). Youth with a low SES have the highest rate of mental illness compared to any other SES group (Reiss et al., 2019). Inequalities between low SES and high SES stand as barriers that not only restrict access to adequate healthcare, but create problems that require more specialized approaches. More data on the biomarkers related to issues that can affect those of a low SES, such as childhood adversity, need to be identified so they can be targeted specifically and become integrated into medical treatment of depression (Torres-Berrío et al., 2019).
It is possible that the stressors of having low SES and having limited access to health care introduces epigenetic changes that cause adverse mental health outcomes. By identifying links between genes associated with mental health and epigenetics, the prevention and treatment techniques for these issues can become more functional for the people affected by a low SES in particular.
Methods
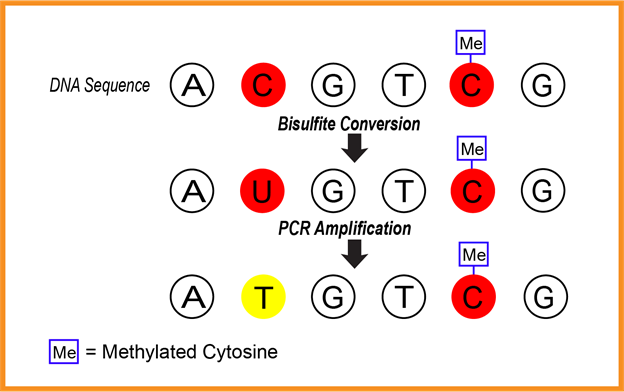
One way of capturing methylation levels is through bisulfite conversion and sequencing. Bisulfite conversion occurs when a strand of DNA is treated with sodium bisulfate. The cytosines that are methylated will remain as cytosines, while those without an extra methyl group will be converted to uracils (see Figure 1). To determine methylation levels, bisulfite sequencing or methylation specific polymerase chain reaction (PCR) is commonly used, which is a method that produces many copies of DNA. Following PCR, which turns the uracils into thymines, the number of cytosine to thymine conversions are counted. The proportion of how many cytosines were converted to thymines and how many were not converted tells us about the extent of the DNA methylation (Wreczycka et al., 2017). The results can reveal the silencing of certain genes due to the addition of an extra methyl group to the cytosines. This method has been applied to genes associated with mental health listed in Table 1 in the results section.
Measuring socioeconomic status is less straightforward. Educational attainment, neighborhood and family income, and occupation are commonly used together or separately to put participants within a sample population into different brackets of SES. Some studies may create their own method for putting their participants in a SES group. For example, there are no universal standards for weighing different levels of educational attainment, resulting in different applications of this factor in studies. Other studies may not use all of the factors associated with SES. In addition to inconsistent research methods, SES itself can change over the lifespan. It is hard to determine SES for people on an individual level, much unlike genomic analysis (Shavers, 2007). In the primary study analyzed in this review, parental educational attainment and family income were used to establish SES groups in their sample. Education level had seven categories, ranging from “less than 9th grade” to graduate school. Income measurements ranged from less than $10,000 to over $150,000 a year, with 8 different levels in between (Swartz et al., 2017).
Measuring socioeconomic status is less straightforward.
Results
| Gene | Conclusions | SES Criteria |
|---|---|---|
|
||
| (Gouin et al., 2017) OXTR | Methylation was only significant on one CpG site in females. |
|
| (Needham et al., 2015) AVP | Methylation present from childhood to adulthood, but upward social mobility lessened the methylation. |
|
|
Serotonin Transporter Gene: SLC6A4
The serotonin transporter gene, SLC6A4, was looked at by two studies. This gene encodes the transporter of serotonin, which is highly important for regulation of mood. Imbalances in serotonin levels are linked to depression. Both studies found that a low SES was associated with more methylation on CpG sites surrounding this gene. The Swartz et al. (2017) study separated the population by whether they had a family history of depression or not, and found that low SES corresponded to more methylation regardless of family history. For those with and without a family history of depression, lower SES at ages 11-15 predicted more methylation at the SLC6A4 proximal promoter. In the Jones-Mason et al. (2016) study on the SLC6A4 gene, unresolved loss and trauma was used as a separate variable from SES. Of the participants that the study considered to have unresolved loss, those with low SES had higher methylation levels on the SLC6A4 gene than their mid to high SES level counterparts. However, low SES did not seem to affect methylation in those without unresolved loss or trauma. Overall, the studies indicate that unresolved loss or trauma is associated with more methylation, but having a family
Oxytocin Receptor Gene: OXTR
The OXTR gene (the oxycontin receptor gene that is often tied to stress response) was targeted by a study (Gouin et al., 2017) that used child abuse as a factor to compare methylation to, in conjunction with SES. The study noted that experiencing a low SES and neglect during childhood was linked to more anxiety throughout life. Childhood anxiety and OXTR gene associations were only significant on one CpG site in females but not males. This only occurred in participants that were put into the “high early life adversity” grouping.
Stress hormones: AVP
In a 2015 study (Needham et al., 2015), methylation of the stress-associated AVP gene was found to be associated with low SES within childhood and adulthood. However, those who had a significantly higher SES as an adult than when they were children had more similarities with their higher SES counterparts. This implied that upward social mobility may “reverse” changes that occurred in the epigenome during childhood, demonstrating plasticity.
Neurological connections: AIFM1, FRY, and LRRN4
A 2019 study (Santos, Jr., et al., 2019) examined SES-related maternal adversity (evaluated by the mother’s marital status and educational attainment) and showed patterns of methylation in genes associated with neurodevelopment using placenta tissue. Two genes were associated with SES adversity, the AIFM1 gene, in which a lack of function could cause neurodegeneration in humans, and the FRY gene, which contributes to healthy neurological development. The authors did note that placenta tissue is still different from other tissues in the body and that the effects beyond this period of development should be further researched. A different study done in 2019 (Laubach et al., 2019) used umbilical blood at birth and found that unusually lower methylation of the LRRN4 gene was associated with low prenatal SES and the low methylation levels persisted through childhood. The LRRN4 gene is important for long-term memories, and its hypomethylation is linked to schizophrenia.
In general, most studies predicted that a low SES would correlate with methylation patterns that disrupt certain genes or biological functions.
...studies predicted that a low SES would correlate with methylation patterns that disrupt certain genes...
Conclusion
Overall, the studies found some correlation between low SES and methylation of different genes. The fluidity of methylation on the AVP gene as participants went from a low SES to a high SES further supports the idea that methylation is linked to SES.
All of the studies used blood or saliva as a source for DNA except for the Santos, Jr. et al. (2019) study which used placental tissue. It was acknowledged that the CpG patterns in the placenta may be difficult to apply to other tissues. Blood cell type proportions (such as the number of natural killer cells and monocytes, which are higher in those with a low SES) and methylation are correlated. It has been argued that the 2017 SLC6A4 promoter study needs to take into account these ratios in order to draw stronger associations with SES (Moore et al., 2020).
The field of epigenomics is still developing and finding its footing in terms of consistency with empirical evidence and taking into account the many different facets of socioeconomic status.
About the Author
Kayla Hartman is a senior at Charles H. Flowers High School in Springdale, Maryland. She is interested in how biological mechanisms affect us psychosocially and is pursuing a career in school psychology.